
We introduce a method to provide vectorial representations of visualĬlassification tasks which can be used to reason about the nature of those Compilation settings are chosen such that the model converges and overfitting is minimized.Task2Vec: Task Embedding for Meta-Learning In case of larger image sizes the batch size is minimal to avoid memory issues. Image sizes are either based on the minimal image size in the dataset, or to avoid long training times or unnecessary resizing. Table III: Image sizes and compilation settings for all source datasets used during pretraining. \printbibliography Appendix A - Pretraining settings -a Pretraining Source Dataset Veronika Cheplygina is supported by the NovoNordisk Foundation (starting package grant) from May 2021, however, the work in this paper was done previously to this award. We thank Colin Nieuwlaat, Felix Schijve and Thijs Kooi for contributing to the discussions about this project. The non-medical datasets ImageNet, STL-10, STI-10, DTD, and the medical datasets PCam-small and KimiaPath960 are used as source dataset only, the other medical datasets ISIC2018, Chest X-rays and PCam-middle are used as source and target dataset. Table I: Datasets used in the experiments with properties name, medical or non-medical origin, size, number of classes, image sizes, and number of channels (RGB/Grey-scale).

ISIC 2018 - Task 3 (Training set) \citepisic2018, tschandl2018ham10000Ĭhest X-rays \citepkermany2018identifying After this, 48 regions of interest of the same size were selected and downsampled to 308x 168 from all 20 WSI scans.ĭescribable Textures Dataset (DTD) \citepcimpoi14describing The twenty classes are based on the image selection process: out of 400 whole slide images of muscle, epithelial and connective tissue 20 images visually representing different pattern types were chosen. The KimiaPath960 classification dataset contains 960 histopathology images of size 308x 168 labeled according to 20 different classes \citepkumar2017comparative. They show this for several applications, although it is worth noting that for natural images, only versions of two related datasets (Tiny ImageNet and CIFAR-10) are examined. They show it is predictive of the transferability of datasets, defined by the relative decrease in error when a source dataset is used.
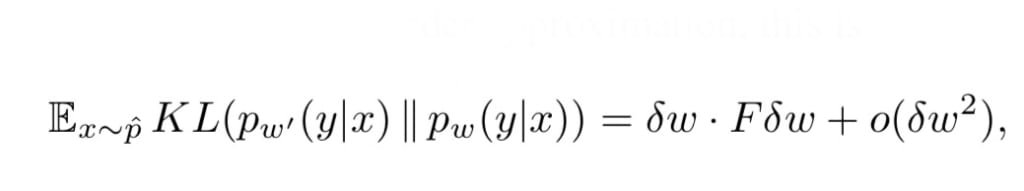
They combine similarity measures based on comparing distributions of words or topics with diversity features based on the source data, and show that diversity combines complementary information.Īnother example of dataset source selection is \citepalvarez2020geometric where the authors propose a distance measure based on the feature-label distributions (i.e., considering both the domain and the task) of the datasets. In an example from natural language processing \citepruder2017learning, the authors learn a model-independent measure of dataset similarity, weighting meta-features using examples from the target dataset. Our experiments and pretrained models are available via \addbibresourceĭataset source selection has been studied for various applications. Finally, we discuss several steps needed for further research in this field, especially with regard to other types (for example 3D) medical images. We show that common intuitions about similarity may be inaccurate, and therefore not sufficient to predict an appropriate source a priori.

We also study different definitions of data similarity. We find that ImageNet is the source leading to the highest performances, but also that larger datasets are not necessarily better. In this paper we perform a systematic study with nine source datasets with natural or medical images, and three target medical datasets, all with 2D images.
TASK2VEC HOW TO
There is currently no consensus on how to choose appropriate source data, and in the literature we can find both evidence of favoring large natural image datasets such as ImageNet, and evidence of favoring more specialized medical datasets. Transfer learning is a commonly used strategy for medical image classification, especially via pretraining on source data and fine-tuning on target data.


 0 kommentar(er)
0 kommentar(er)
